Example ML Flow
Introduction
We’ll consider a typical (but simplified) machine learning flow.
(The code for this example is available in the Bionic repo at
example/ml_workflow.py. Run pip install 'bionic[examples]'
to ensure you have the required dependencies.)
1"""
2A toy ML workflow intended to demonstrate basic Bionic features. Trains a
3logistic regression model on the UCI ML Breast Cancer Wisconsin (Diagnostic)
4dataset.
5"""
6
7import re
8
9import pandas as pd
10from sklearn import datasets, linear_model, metrics, model_selection
11
12import bionic as bn
13
14# Initialize our builder.
15builder = bn.FlowBuilder("ml_workflow")
16
17# Define some basic parameters.
18builder.assign(
19 "random_seed", 0, doc="Arbitrary seed for all random decisions in the flow."
20)
21builder.assign(
22 "test_split_fraction", 0.3, doc="Fraction of data to include in test set."
23)
24builder.assign(
25 "hyperparams_dict", {"C": 1}, doc="Hyperparameters to use when training the model."
26)
27builder.assign(
28 "feature_inclusion_regex",
29 ".*",
30 doc="Regular expression specifying which feature names to include.",
31)
32
33
34# Load the raw data.
35@builder
36def raw_frame():
37 """
38 The raw data, including all features and a `target` column of labels.
39 """
40 dataset = datasets.load_breast_cancer()
41 df = pd.DataFrame(data=dataset.data, columns=dataset.feature_names)
42 df["target"] = dataset.target
43 return df
44
45
46# Select a subset of the columns to use as features.
47@builder
48def features_frame(raw_frame, feature_inclusion_regex):
49 """Labeled data with a selected subset of the feature columns."""
50 included_feature_cols = [
51 col
52 for col in raw_frame.columns.drop("target")
53 if re.match(feature_inclusion_regex, col)
54 ]
55 return raw_frame[included_feature_cols + ["target"]]
56
57
58# Split the data into train and test sets.
59@builder
60# The `@outputs` decorator tells Bionic to define two new entities from this
61# function (which returns a tuple of two values).
62@bn.outputs("train_frame", "test_frame")
63@bn.docs(
64 "Subset of feature data rows, used for model training.",
65 "Subset of feature data rows, used for model testing.",
66)
67def split_raw_frame(features_frame, test_split_fraction, random_seed):
68 return model_selection.train_test_split(
69 features_frame,
70 test_size=test_split_fraction,
71 random_state=random_seed,
72 )
73
74
75# Fit a logistic regression model on the training data.
76@builder
77def model(train_frame, random_seed, hyperparams_dict):
78 """A binary classifier sklearn model."""
79 m = linear_model.LogisticRegression(
80 solver="liblinear", random_state=random_seed, **hyperparams_dict
81 )
82 m.fit(train_frame.drop("target", axis=1), train_frame["target"])
83 return m
84
85
86# Predict probabilities for the test data.
87@builder
88def prediction_frame(model, test_frame):
89 """
90 A dataframe with one column, `proba`, containing predicted probabilities for the
91 test data.
92 """
93 predictions = model.predict_proba(test_frame.drop("target", axis=1))[:, 1]
94 df = pd.DataFrame()
95 df["proba"] = predictions
96 return df
97
98
99# Evaluate the model's precision and recall over a range of threshold values.
100@builder
101def precision_recall_frame(test_frame, prediction_frame):
102 """
103 A dataframe with three columns:
104 - `threshold`: a probability threshold for the model
105 - `precision`: the test set precision resulting from that threshold
106 - `recall`: the test set recall resulting from that threshold
107 """
108 precisions, recalls, thresholds = metrics.precision_recall_curve(
109 test_frame["target"],
110 prediction_frame["proba"],
111 )
112
113 df = pd.DataFrame()
114 df["threshold"] = [0] + list(thresholds) + [1]
115 df["precision"] = list(precisions) + [1]
116 df["recall"] = list(recalls) + [0]
117
118 return df
119
120
121# Plot the precision against the recall.
122@builder
123# The `@pyplot` decorator makes the Matplotlib plotting context available to
124# our function, then translates our plot into an image object.
125@bn.pyplot("plt")
126# The `@gather` decorator collects the values of of "hyperparams_dict" and
127# "precision_recall_frame" into a single dataframe named "gathered_frame".
128# This might not seem very interesting since "gathered_frame" only has one row,
129# but it will become useful once we introduce multiplicity.
130@bn.gather(
131 over="hyperparams_dict", also="precision_recall_frame", into="gathered_frame"
132)
133def all_hyperparams_pr_plot(gathered_frame, plt):
134 """
135 A plot of precision against recall. Includes one curve for each set of
136 hyperparameters.
137 """
138 _, ax = plt.subplots(figsize=(4, 3))
139 for row in gathered_frame.itertuples():
140 label = ", ".join(
141 f"{key}={value}" for key, value in row.hyperparams_dict.items()
142 )
143 row.precision_recall_frame.plot(x="recall", y="precision", label=label, ax=ax)
144 ax.set_xlabel("Recall")
145 ax.set_ylabel("Precision")
146
147
148# Assemble our flow object.
149flow = builder.build()
Let’s start by importing the flow into a notebook and visualizing it.
[2]:
from example.ml_workflow import flow
flow.render_dag()
[2]:
This is still a toy example, but it demonstrates a classic supervised machine learning flow for binary classification:
Load data (
raw_frame)Clean it (
features_frame)Split into training and test sets (
train_frameandtest_frame)Fit a model on the training data (
model)Evaluate the model on the test data (
predictions_frame,precision_recall_frame)Visualize the model’s performance (
all_hyperparams_pr_plot)
As in the previous example, we can access any of the entity values. However, this time we’ll enable logging so we can see every entity that Bionic computes.
[3]:
import logging; logging.basicConfig(level='INFO', format='%(message)s')
flow.get.train_frame().head()
Accessed random_seed(random_seed=0) from definition
Accessed test_split_fraction(test_split_fraction=0.3) from definition
Accessed feature_inclusion_regex(feature_inclusion_regex=_.._12aca9f6a2) from definition
Computing raw_frame() ...
Computed raw_frame()
Computing features_frame(feature_inclusion_regex=_.._12aca9f6a2) ...
Computed features_frame(feature_inclusion_regex=_.._12aca9f6a2)
Computing (train_frame, test_frame)(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) ...
Computed (train_frame, test_frame)(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0)
Computing (train_frame, test_frame)(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) ...
Computed (train_frame, test_frame)(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0)
[3]:
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 478 | 11.490 | 14.59 | 73.99 | 404.9 | 0.10460 | 0.08228 | 0.05308 | 0.01969 | 0.1779 | 0.06574 | ... | 21.90 | 82.04 | 467.6 | 0.1352 | 0.2010 | 0.25960 | 0.07431 | 0.2941 | 0.09180 | 1 |
| 303 | 10.490 | 18.61 | 66.86 | 334.3 | 0.10680 | 0.06678 | 0.02297 | 0.01780 | 0.1482 | 0.06600 | ... | 24.54 | 70.76 | 375.4 | 0.1413 | 0.1044 | 0.08423 | 0.06528 | 0.2213 | 0.07842 | 1 |
| 155 | 12.250 | 17.94 | 78.27 | 460.3 | 0.08654 | 0.06679 | 0.03885 | 0.02331 | 0.1970 | 0.06228 | ... | 25.22 | 86.60 | 564.2 | 0.1217 | 0.1788 | 0.19430 | 0.08211 | 0.3113 | 0.08132 | 1 |
| 186 | 18.310 | 18.58 | 118.60 | 1041.0 | 0.08588 | 0.08468 | 0.08169 | 0.05814 | 0.1621 | 0.05425 | ... | 26.36 | 139.20 | 1410.0 | 0.1234 | 0.2445 | 0.35380 | 0.15710 | 0.3206 | 0.06938 | 0 |
| 101 | 6.981 | 13.43 | 43.79 | 143.5 | 0.11700 | 0.07568 | 0.00000 | 0.00000 | 0.1930 | 0.07818 | ... | 19.54 | 50.41 | 185.2 | 0.1584 | 0.1202 | 0.00000 | 0.00000 | 0.2932 | 0.09382 | 1 |
5 rows × 31 columns
[4]:
flow.get.all_hyperparams_pr_plot()
Accessed hyperparams_dict(hyperparams_dict=49b64ab8bc) from definition
Accessed train_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) from in-memory cache
Accessed random_seed(random_seed=0) from in-memory cache
Computing model(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc) ...
Computed model(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc)
Loaded test_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) from disk cache
Computing prediction_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc) ...
Computed prediction_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc)
Computing precision_recall_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc) ...
Computed precision_recall_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc)
Computing all_hyperparams_pr_plot(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) ...
generated new fontManager
Computed all_hyperparams_pr_plot(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0)
[4]:
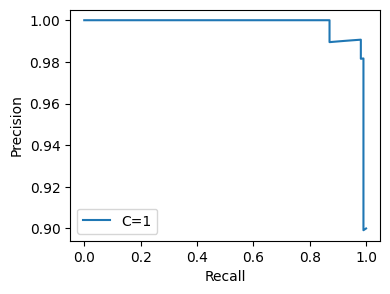
<Figure size 640x480 with 0 Axes>
Note that in the logs, each entity is computed exactly once – every value is cached both in memory and on disk to avoid redundant computation.
Changing Inputs
Now that we have our flow, we might want to experiment with
different parameter settings.
For example, we can try changing our regularization hyperparameter C:
[5]:
flow.setting('hyperparams_dict', {'C': 10}).get.all_hyperparams_pr_plot()
Accessed hyperparams_dict(hyperparams_dict=dcec991282) from definition
Loaded train_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) from disk cache
Loaded random_seed(random_seed=0) from disk cache
Computing model(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=dcec991282) ...
Computed model(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=dcec991282)
Loaded test_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) from disk cache
Computing prediction_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=dcec991282) ...
Computed prediction_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=dcec991282)
Computing precision_recall_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=dcec991282) ...
Computed precision_recall_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=dcec991282)
Computing all_hyperparams_pr_plot(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) ...
Computed all_hyperparams_pr_plot(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0)
[5]:
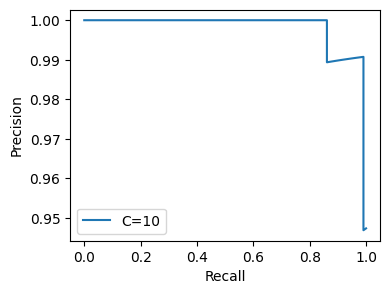
<Figure size 640x480 with 0 Axes>
Bionic re-runs the flow with the changed parameter, generating a new plot.
Since we’ve created a new copy of our flow, Bionic doesn’t try to reuse its in-memory cache. However, we can see it that it only recomputed those entities whose values changed – the others were loaded from disk.
Similarly, we can try training our model on a subset of the available features:
[6]:
flow.setting('feature_inclusion_regex', 'mean.*').get.all_hyperparams_pr_plot()
Accessed feature_inclusion_regex(feature_inclusion_regex=mean.._a0ed126bd6) from definition
Loaded raw_frame() from disk cache
Computing features_frame(feature_inclusion_regex=mean.._a0ed126bd6) ...
Computed features_frame(feature_inclusion_regex=mean.._a0ed126bd6)
Loaded test_split_fraction(test_split_fraction=0.3) from disk cache
Loaded random_seed(random_seed=0) from disk cache
Computing (train_frame, test_frame)(feature_inclusion_regex=mean.._a0ed126bd6, test_split_fraction=0.3, random_seed=0) ...
Computed (train_frame, test_frame)(feature_inclusion_regex=mean.._a0ed126bd6, test_split_fraction=0.3, random_seed=0)
Computing (train_frame, test_frame)(feature_inclusion_regex=mean.._a0ed126bd6, test_split_fraction=0.3, random_seed=0) ...
Computed (train_frame, test_frame)(feature_inclusion_regex=mean.._a0ed126bd6, test_split_fraction=0.3, random_seed=0)
Loaded hyperparams_dict(hyperparams_dict=49b64ab8bc) from disk cache
Computing model(feature_inclusion_regex=mean.._a0ed126bd6, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc) ...
Computed model(feature_inclusion_regex=mean.._a0ed126bd6, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc)
Computing prediction_frame(feature_inclusion_regex=mean.._a0ed126bd6, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc) ...
Computed prediction_frame(feature_inclusion_regex=mean.._a0ed126bd6, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc)
Computing precision_recall_frame(feature_inclusion_regex=mean.._a0ed126bd6, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc) ...
Computed precision_recall_frame(feature_inclusion_regex=mean.._a0ed126bd6, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc)
Computing all_hyperparams_pr_plot(feature_inclusion_regex=mean.._a0ed126bd6, test_split_fraction=0.3, random_seed=0) ...
Computed all_hyperparams_pr_plot(feature_inclusion_regex=mean.._a0ed126bd6, test_split_fraction=0.3, random_seed=0)
[6]:
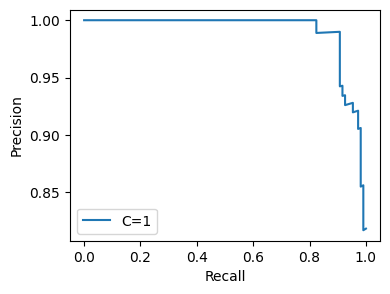
<Figure size 640x480 with 0 Axes>
Naturally, reducing the number of features makes the model perform worse.
We can also apply this flow to a completely different dataset, such as the UCI ML Wine dataset. (This is actually a multiclass dataset, but to match the original example we’ll use binary labels.)
[7]:
from sklearn import datasets
import pandas as pd
wine_data = datasets.load_wine()
wine_frame = pd.DataFrame(
data=wine_data['data'],
columns=wine_data['feature_names'])
wine_frame['target'] = (wine_data['target'] == 0)
wine_flow = flow.setting('raw_frame', wine_frame)
wine_flow.get.all_hyperparams_pr_plot()
Accessed raw_frame(raw_frame=40e4b932f4) from definition
Loaded feature_inclusion_regex(feature_inclusion_regex=_.._12aca9f6a2) from disk cache
Computing features_frame(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2) ...
Computed features_frame(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2)
Loaded test_split_fraction(test_split_fraction=0.3) from disk cache
Loaded random_seed(random_seed=0) from disk cache
Computing (train_frame, test_frame)(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) ...
Computed (train_frame, test_frame)(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0)
Computing (train_frame, test_frame)(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) ...
Computed (train_frame, test_frame)(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0)
Loaded hyperparams_dict(hyperparams_dict=49b64ab8bc) from disk cache
Computing model(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc) ...
Computed model(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc)
Computing prediction_frame(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc) ...
Computed prediction_frame(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc)
Computing precision_recall_frame(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc) ...
Computed precision_recall_frame(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc)
Computing all_hyperparams_pr_plot(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) ...
Computed all_hyperparams_pr_plot(raw_frame=40e4b932f4, feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0)
[7]:
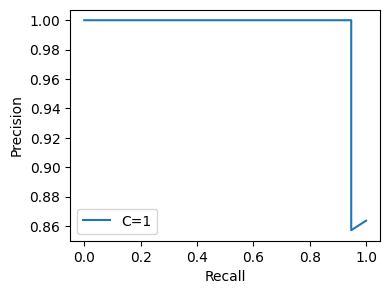
<Figure size 640x480 with 0 Axes>
Multiplicity
Now we have a flow for evaluating any given set of parameters. But usually we want to compare multiple variations at once, within a single flow. Bionic lets us do this by setting multiple values for a single entity:
[8]:
flow3 = flow.setting('hyperparams_dict', values=[{'C': 0.1}, {'C': 1}, {'C': 10}])
flow3.get.all_hyperparams_pr_plot()
Accessed hyperparams_dict(hyperparams_dict=516eeb1664) from definition
Loaded train_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) from disk cache
Loaded random_seed(random_seed=0) from disk cache
Computing model(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=516eeb1664) ...
Computed model(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=516eeb1664)
Loaded test_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) from disk cache
Computing prediction_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=516eeb1664) ...
Computed prediction_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=516eeb1664)
Computing precision_recall_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=516eeb1664) ...
Computed precision_recall_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=516eeb1664)
Loaded hyperparams_dict(hyperparams_dict=49b64ab8bc) from disk cache
Loaded precision_recall_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc) from disk cache
Loaded precision_recall_frame(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=dcec991282) from disk cache
Loaded hyperparams_dict(hyperparams_dict=dcec991282) from disk cache
Computing all_hyperparams_pr_plot(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0) ...
Computed all_hyperparams_pr_plot(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0)
[8]:
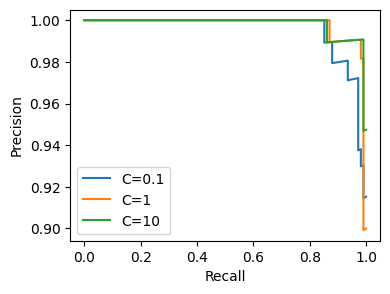
<Figure size 640x480 with 0 Axes>
Here we can see the PR curves for each of three hyperparameter configurations. But how did this work? Let’s start by visualizing the dependency graph of our new flow:
[9]:
flow3.render_dag()
Accessed core__flow_name(core__flow_name=ml_workflow) from in-memory cache
[9]:
When we specify multiple values for an entity,
Bionic creates multiple instances of that entity and of all downstream entities.
If there are three instances of hyperparams_dict,
there must also be three instances of model and precision_recall_frame.
However, we can also see that there is only one instance of all_hyperparams_plot_pr.
This is because we used the @gather decorator when defining this entity:
Bionic has aggregated all the variations of hyperparameters_dict and precision_recall_frame
into a single frame, so they can be plotted together.
Now, because we have three different model instances, it no longer makes sense to request “the” model:
[10]:
try:
flow3.get.model()
except Exception as e:
caught_exception = e
caught_exception
Accessed model(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=516eeb1664) from in-memory cache
Loaded model(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc) from disk cache
Loaded model(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=dcec991282) from disk cache
[10]:
ValueError("Entity 'model' has multiple values")
If we want to access an entity with multiple values, we need to tell Bionic what data structure to use to aggregate them:
[11]:
flow3.get.model('series')
Accessed model(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=516eeb1664) from in-memory cache
Accessed model(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=49b64ab8bc) from in-memory cache
Accessed model(feature_inclusion_regex=_.._12aca9f6a2, test_split_fraction=0.3, random_seed=0, hyperparams_dict=dcec991282) from in-memory cache
Loaded feature_inclusion_regex(feature_inclusion_regex=_.._12aca9f6a2) from disk cache
Accessed hyperparams_dict(hyperparams_dict=516eeb1664) from in-memory cache
Accessed hyperparams_dict(hyperparams_dict=49b64ab8bc) from in-memory cache
Accessed hyperparams_dict(hyperparams_dict=dcec991282) from in-memory cache
Accessed random_seed(random_seed=0) from in-memory cache
Loaded test_split_fraction(test_split_fraction=0.3) from disk cache
[11]:
feature_inclusion_regex hyperparams_dict random_seed test_split_fraction
W(.*) W({'C': 0.1}) W(0) W(0.3) LogisticRegression(C=0.1, random_state=0, solv...
W({'C': 1}) W(0) W(0.3) LogisticRegression(C=1, random_state=0, solver...
W({'C': 10}) W(0) W(0.3) LogisticRegression(C=10, random_state=0, solve...
Name: model, dtype: object
Extended Multiplicity
We can apply the same approach to any number of entities:
[12]:
flow6 = flow3.setting('feature_inclusion_regex', values=['.*', 'mean.*'])
flow6.render_dag()
Accessed core__flow_name(core__flow_name=ml_workflow) from in-memory cache
[12]:
Here we can see that each entity’s multiplicity is propagated through the entire graph.
For example, there are two train_frame nodes and two test_frame nodes,
because those depend on feature_inclusion_regex but not on hyperparams_dict.
However, there are six model nodes, because there are six (two times three) combinations of
train_frame and hyperparams_dict.
We can also see two instances of all_hyperparams_pr_plot,
corresponding to the two instances of feature_inclusion_regex.
This illustrates another nuance of how we used @gather:
we asked Bionic to include every variation of hyperparams_dict,
along with the associated values of precision_recall_frame.
If we had used
@gather(
over=['hyperparams_dict', 'feature_inclusion_regex'],
also='precision_recall_frame',
into='gathered_frame')
def all_variations_pr_plot(...):
...
then all the values would be gathered into a single frame, and there would be one single plot node.
Wrapping Up
This tutorial has illustrated four topics:
How to use Bionic to assemble a more complex data science flow.
How Bionic uses caching to avoid redundant computation.
How to repurpose an existing flow to use new parameters or data.
How to use multiplicity to compare several variations within a single flow.
These concepts should be enough to get started building real flows. The next section of the documentation will discuss the underlying concepts in Bionic.